# Load the package:
library(MVN)Multivariate Outlier Detection
Before interpreting your multivariate normality tests, it’s important to check for and understand any influential outliers. In this section, we’ll:
- Detect multivariate outliers using robust Mahalanobis distances.
- Summarize flagged observations via the summary method.
- Visualize outliers in Q–Q and scatter plots.
Example Data
We’ll use two numeric variables from the built-in iris dataset:
df <- iris[1:50, 1:2]
head(df) Sepal.Length Sepal.Width
1 5.1 3.5
2 4.9 3.0
3 4.7 3.2
4 4.6 3.1
5 5.0 3.6
6 5.4 3.91. Detecting Outliers
The mvn() function can automatically flag multivariate outliers using methods such as the adjusted quantile approach ("adj") or a fixed quantile cutoff. Specify via multivariate_outlier_method:
out_res <- mvn(
data = df,
mvn_test = "hz",
multivariate_outlier_method = "quan"
)This computes robust Mahalanobis distances and flags observations above the chi-square cutoff at the specified alpha (default 0.05).
2. Summarizing Outliers
Use the summary() function with select = "outliers" to list flagged observations:
summary(out_res, select = "outliers")── Multivariate Outliers ─────────────────────────────────────────────────────── Observation Mahalanobis.Distance
1 15 10.700
2 42 10.263
3 14 9.675
4 19 9.174
5 16 9.076
6 23 8.742
7 43 8.710The output shows each outlier’s observation index and Mahalanobis distance, helping you decide whether to inspect or remove these points.
3. Visualizing Outliers
plot(out_res, diagnostic = "outlier")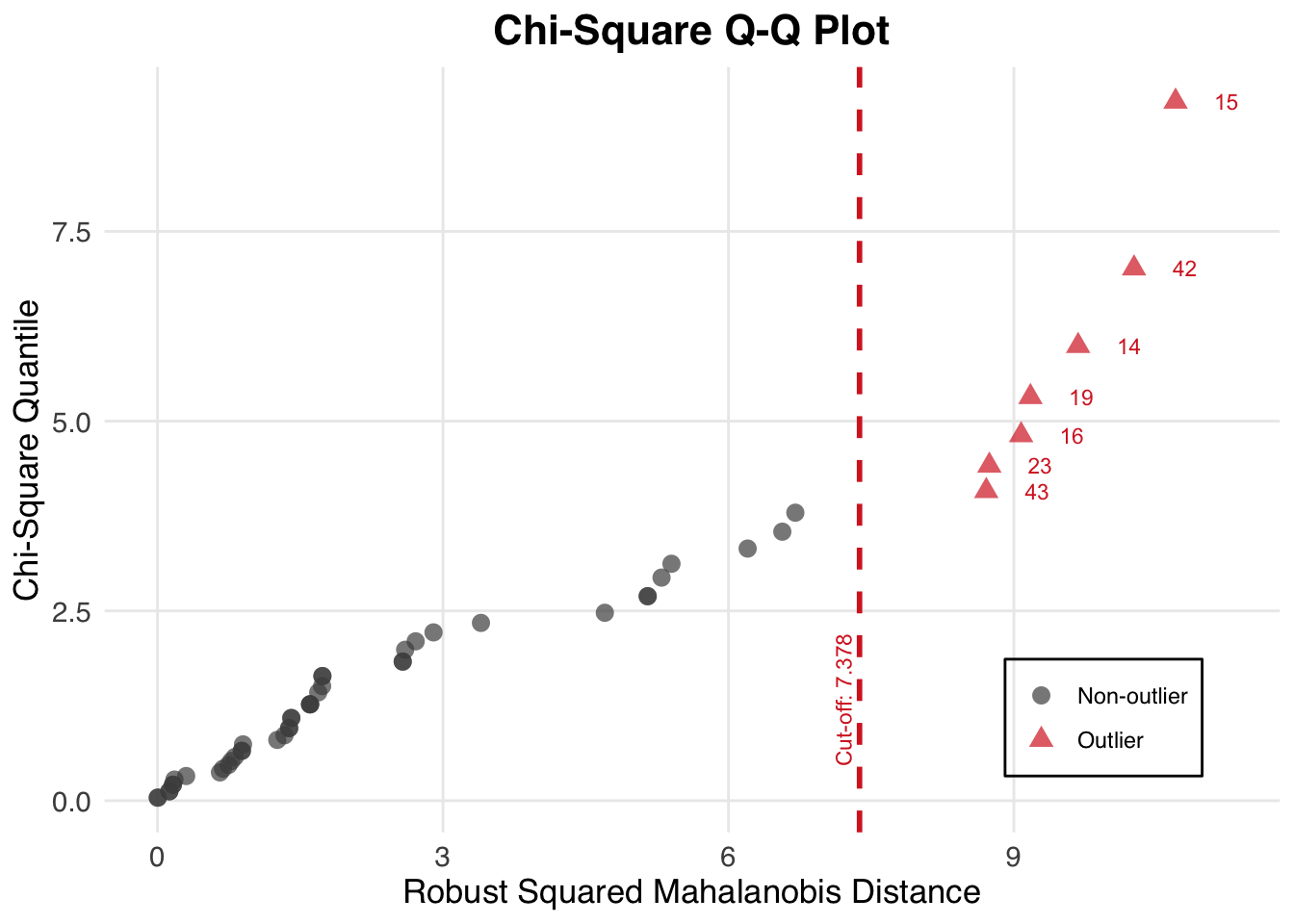
This Q–Q plot highlights points deviating from the theoretical chi-square line.
References
Korkmaz S, Goksuluk D, Zararsiz G. MVN: An R Package for Assessing Multivariate Normality. The R Journal. 2014;6(2):151–162. URL: https://journal.r-project.org/archive/2014-2/korkmaz-goksuluk-zararsiz.pdf